|
Assessing
Molecular
Pathogenicity for
Rare
Diseases
|
|
|
According to the specific aims we defined two ELIXIR fields of activity for:
For the practical implementation we collected a set of resources and tools already included in the ELIXIR bio.tools registry for addressing each task. In the first activity (Fig. 1) we focus initially on the annotation and interpretation of the small variants (SNV and indels) for identifying a set of “actionable” variants. In this context, we defined a specific procedures for predicting the impact of the genetic variants at protein level using structural and functional information (Fig. 2). |
From DNAseq data to disease report (version 0.1)
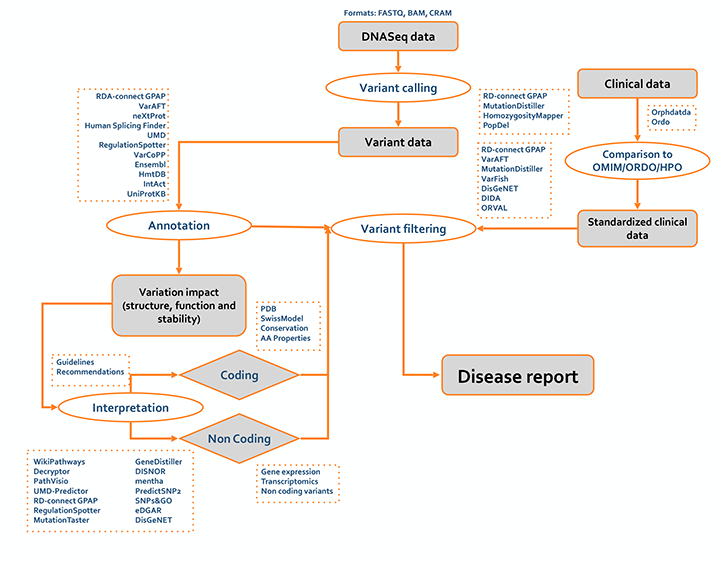
|
Predict the impact of protein variants (version 0.1)
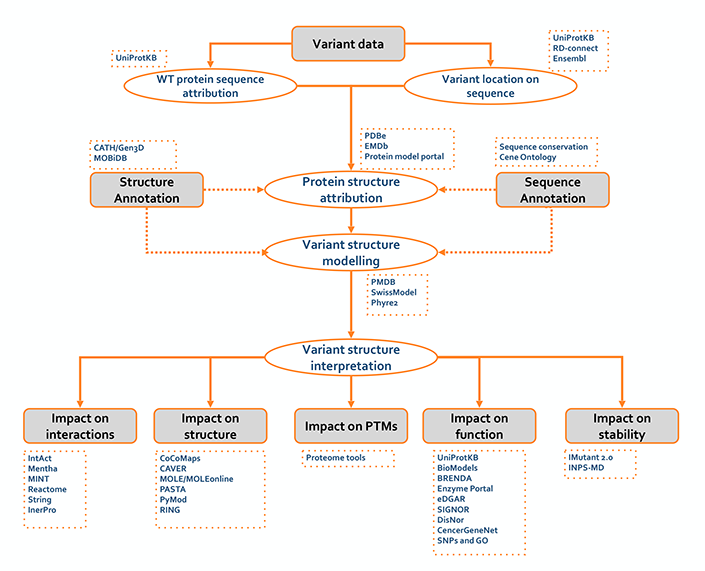
|
|
The above flowcharts are subject to changes as the working group advances. In the next versions, the workflows will be updated including references to tools, publications and bioinformatics resources. Furthermore our team is collecting a pool of ELIXIR and external training activities to familiarize scientists from different research fields with the state-of-the-art resource and tools for variant interpretation. |
